Input Script@
- This is a
REQUIREDfile. - This file provides the simulation parameters and the filenames of other required inputs required to carry out irradiation simulations, and the output filenames
- Lines starting with
#(hash symbols) are considered as comments.- Each line has to be commented individually with
#. - There is NO option to comment multiple lines simultaneously.
- Each line has to be commented individually with
- Syntax coloring of keywords is available for emacs and textmate editors
- All keywords must be in the
lowercase - NOTE: Keywords marked as
OPTIONALdoes not mean that they are not required, but default values will be used if the user does provide them.
List of Keywords by Category@
Related to Input Files@
The following keywords are used to specify the names of input files for carrying out irradiation simulations. Including the input script file, there are 7 input files. Of which the file read using the keyword read_extd_func is an optional file in all cases.
Except for the keyword read_cascade, Usage is keyword <filename>
| Keywords | Purpose | Required |
|---|---|---|
diff_db | Filename for activated Event DB | YES |
react_db | Filename for Reaction Event DB | YES |
read_data | Filename for Initial State of the system. Usage see description | YES if cascade_dep_rate ≠ 0Otherwise NOT RELEVANT |
read_extd_func | Filename for analytical expressions for extended defects | YES |
cap_radius_db | Filename for capture radii of defect complexes when analytical forms DO NOT exist | YES |
read_cascade | Filenames of cascades (entire name except the number) Usage: see description | YES |
read_cprad_func | Filename for analytical expressions for capture radii of defect complexes | OPTIONAL |
Related to Output Files@
- Following keywords provide the filenames where the output data will be saved at the end of the simulation.
- Use of these keywords is not required if one uses default filenames.
| Keywords | Purpose | Required |
|---|---|---|
dumpconfig | Filename for saving snap shots Usage: see description | OPTIONAL |
react_count | Filename for saving reaction event counts as a function of simulation time Usage: react_count <filename> Default: react_count reaction_pcount_Vs_time | OPTIONAL |
ty_sz_time | Prefix for the filename for saving defect count as a function of time and size. This information for each defect type is saved in separate files with a filename starting with the prefix. Usage: ty_sz_time <fname_prefix>(also see description) | OPTIONAL |
bcrossings_sz_time | Prefix for the filename for saving defect counts as a function of time and size that cross the simulation box with absorbing boundary. Usage: bcrossings_sz_time <fname_prefix> (also see description) | OPTIONAL |
dumpdensities | Filename for saving the data on defect densities. Usage: dumpdensities <filename> Default: dumpdensitites densitites_Vs_time | OPTIONAL |
bcrossing | Filename where data on the number of defects crossing an absorbing boundary is saved. Usage: see description | OPTIONAL |
max_data_clstr_size | Largest defect cluster size upto which size dependent densities are collected. Usage: max_data_clstr_size <integer values>(also see description) | OPTIONAL |
Note
Data saved in the files associated with the keywords dumpconfig, react_count, bcrossings_sz_time and bcrossing can be collected only during the run-time. While data saved in the files associated with the keywords ty_sz_time and dumpdensities can be obtained by post-processing.
Related to Simulation Box Setup@
| Keywords | Purpose | Required |
|---|---|---|
lattice | Specify lattice Usage: lattice <type> Types: FCC, BCC CUSTOM(also see description) | YES |
directions | Specify Direction vectors Default: None Usage: see description | YES if lattice CUSTOM otherwise NOT RELEVANT |
grain_size | Grain Size (in lattice units) Usage: grain_size <value> (also see description) | YES for finite boundary conditionsOtherwise NOT RELEVANT |
cell_size | Simulation box is divided into following cell sizes Usage: see description | YES |
latt_const | Lattice Constant (in Å) Usage: latt_const <value> | OPTIONAL |
boundary | Specify boundary conditions. Usage: see description Default: PPP (periodic along xyz-directions) | OPTIONAL |
Related to Simulation Conditions@
| Keywords | Purpose | Required |
|---|---|---|
ndata | Number of times data is collected during a simulation. Data is collected at equal intervals of time. Usage: ndata <integer value> | YES |
temperature | Temperature of the simulation (in Kelvin) Usage: temperature <value> | YES |
nsteps | Maximum number KMC steps. Once this number is reached, the simulation will stop Includes DIFFUSION, EMISSION, DEPOSITION and TRANSFORMATION events Usage: nsteps <integer value> | YES |
max_active_clstr | Cluster sizes larger than those specified are not included in rate tables. Usage: see description | YES |
tot_simtime | Max. length of simulation time in seconds Usage: tot_simtime <value> | YES |
defect_names | Provide defect names. These names will be used in output files and command-line output. Usage: see description | OPTIONAL |
emission_radius | Additional radius (in lattice units) added to the sum of capture radii of the parent and emitted defects Default: 2.0Usage: emission_radius <value> (also see description) | OPTIONAL |
seed | Seed for the random number generator (should be an odd number) Usage seed <odd integer value> | YES |
cascade_dep_rate | Cascade deposition rate in cascades/second Default 0Usage: cascade_dep_rate <value>(also see description) | OPTIONAL |
rotate | This turns on random rotation of cascades otherwise cascades are simply translated in space Default: off Usage: rotate <ON or OFF> | OPTIONAL |
max_r_itr | Maximum number of iterations allowed to carry out reaction events recursively Usage: max_r_itr <integer value> Default: 5(also see description) | OPTIONAL |
shift_cntr | Shifts a cascade inserted at a random location to the middle of the simulation cell (only used during single cascade annealing) Default: offUsage: shift_cntr <yes or no>(also see description) | OPTIONAL |
list_size | Increases the size of the matrix that stores defect information Default: 1000 Usage: list_size <value>(also see description) | OPTIONAL |
rcutoff | Cutoff radius for reaction event search (in lattice units) Usage: rcutoff <value> | YES |
rate_scaling | OPTIONAL | |
max_walltime | Maximum wall-clocktime (in seconds) a simulation would run. Usage: max_walltime <value> Default: 3600seconds (also see description) | OPTIONAL |
Description of Keywords@
lattice@
lattice keyword specifies the type of the lattice, thereby the directions of diffusion hops, which by default are chosen to be along the nearest-neighbor directions.
Only the FCC and BCC lattice vectors are hardwired. For other lattice type the option CUSTOM have to be using in conjunction with the keyword directions.
directions@
This keyword is used to define the possible directions of diffusion hops when lattice CUSTOM
Example
In a BCC lattice, if some defect clusters diffuse in BCC directions while other defects in [100] directions, then the hopping directions are inputted as shown below:
USAGE:
lattice CUSTOM
directions 48 1 1 1 1 -1 1 -1 -1 1 -1 1 1 1 1 -1 1 -1 -1 -1 -1 -1 -1 1 -1 1 0 0 -1 0 0 0 1 0 0 -1 0 0 0 -1 0 0 1 0 0 -2 0 0 -3
Number 48 after directions represents the total number of entries. In the present case it is \(\small 8(\text{BCC}) \times 3 + 6 (\text{FCC}) \times 3 + 2 (\text{Multi-hop}) \times 3= 48\) entries.
BCC Directions: 0 (1,1,1) 1 (1,-1,1) 2 (-1,-1,1) 3 (-1,1,1) 4 (1,1,-1) 5 (1,-1,-1) 6 (-1,-1,-1) 7 (-1,1,-1)
[100] Directions: 8 (1,0,0) 9 (-1,0,0) 10 (0,1,0) 11 (0,-1,0) 12 (0,0,-1) 13 (0,0,1)
Multi-Step Hops: 14 (0,0,-2) 15 (0,0,-3)
Constraints
- This option can be only used along with
lattice CUSTOM. - For
lattice BCCorlattice FCC, this keyword is ignored. - The number of entries for
directionsshould be perfectly divisible by3.
boundary@
This keyword specifies how the simulation box boundaries are treated when a defect or an object crosses it. In the present version of the KSOME, three different boundary conditions are allowed. - Absorbing (A) - Periodic (P) - Finite Periodic (F)
Example
boundary PPP\(\Rightarrow\) Periodic Boundary Conditions (PBC) are applied to the simulation box boundaries along xyz-directionsboundary AAA\(\Rightarrow\) Absorbing Boundary Conditions (ABC) are applied to the simulation box boundaries along xyz-directionsboundary FFF\(\Rightarrow\) Finite Boundary Conditions (FBC) are applied to the simulation box boundaries along xyz-directions
One can also apply different boundary conditions on the simulation box boundaries along different directions.
Different Boundary Conditions along xyz-directions
boundary PPA\(\Rightarrow\) PBC is applied on the simulation box boundaries along xy-directions and ABC along the z-directionboundary PFA\(\Rightarrow\) PBC, FBC and ABC along x-, y- and z-directions
For additional information on boundary conditions see kwiki
grain_size@
This option lets the user specify the grain size in terms of lattice units (lu).
Example
In a simulation box with BCC lattice, a lattice unit corresponds to half the lattice constant (a0) . Hence a grain size of 1.0\(\mu\)m corresponds to 6300 lu when \(a_0\) = 3.16 Å. Entry for this item in the input script file would be grain_size 6300
Constraints
- Grain size effect is only applied only on the simulation box boundary with the boundary condition
F(FBC) - Grain size has no effect when periodic boundary conditions are used i.e., when
boundary PPP - To impose the grain size effect on a simulation,
boundary FFFshould be used. - When
boundary FFF, then the simulation box is assumed to be at the center of a spherical grain.
defect_names@
This keyword is used to specify defect names. These names will when printing defect counts to the screen and also in the output files.
Usage: defect_names <num. of defect types> <list of defect names>
Example
defect_names 5 V SIA He HeV HeI
max_active_clstr@
Cluster sizes larger than specified values are not included in rate tables. The usage is similar to defect_names
Usage: max_active_clstr <num. of defect types> <defect cluster size>
Example
defect_names 5 V SIA He HeV HeI max_active_clstr 5 4 200 0 0 0
In this example, vacancy clusters and SIA clusters larger than size 4 and 300, respectively, are inactive, i.e., they neither diffuse nor dissolve via emission. In the case of He, HeV and HeI defect type clusters, all sizes are considered as active.
Warning
This information over-rides any information regarding the diffusion and emission events of a defect cluster in the activated event database.
max_data_clstr_size@
cell_size@
- This
keywordis used to split the simulation cell into smaller boxes to limit the search space for reaction events.
Example
In figures (a) and (b), a simulation box with dimensions \(60 \times 60 \times 60\) is split into 27 smaller boxes with dimensions \(20 \times 20 \times 20\) (Units: lattice units)
USAGE: cell_size 20 20 20
Sphere represents the capture radius of the point defect | |
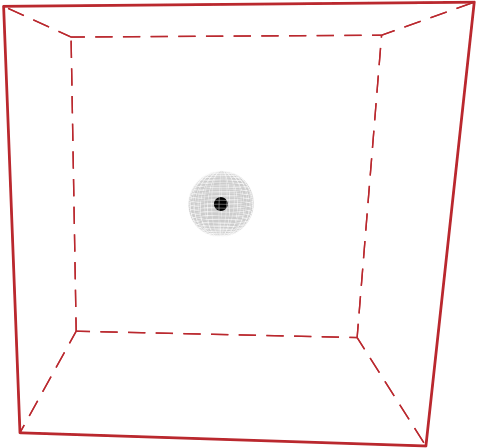 | 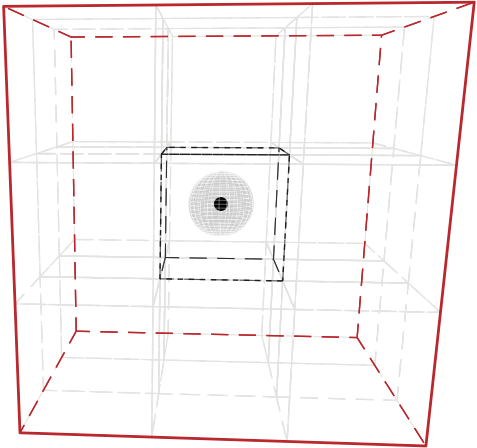 |
Fig: 2D view; Box 4 (blue box) includes defects from the neighboring boxes that are in the shaded region.
Warning
Simulation box dimensions and the number of cells along xyz-directions should be EXACTLY DIVISIBLE.
emission_radius@
Default emission_radius is 2.0, which for a BCC and FCC lattices is equal to one lattice constant (a0).
$$ \usetikzlibrary{arrows, arrows.meta} \usetikzlibrary{positioning,shadows} \usetikzlibrary{backgrounds} \definecolor{wrwrwr}{rgb}{0.3803921568627451,0.3803921568627451,0.3803921568627451} \definecolor{rvwvcq}{rgb}{0.08235294117647059,0.396078431372549,0.7529411764705882} \begin{tikzpicture}[line cap=round,line join=round,>=latex,x=1cm,y=1cm] \scriptsize \draw [line width=1pt,dotted,color=wrwrwr] (-1.9127272727272742,3.5345454545454564) circle (1.19345cm); \draw [->,>=latex,line width=1pt,color=wrwrwr] (-1.9127272727272742,3.5345454545454564) -- (-1.010277758642073,2.753579528894802); \draw [line width=1pt,dotted,color=wrwrwr] (-6.749090909090913,3.534545454545457) circle (2.050921985775066cm); \draw [->,>=latex,line width=1pt,color=wrwrwr] (-6.749090909090913,3.534545454545457) -- (-5.472612873213186,1.9292776215477105); \draw (-4.670447268187056,1.5655281261130782) node[anchor=north west] {Capture Radius}; \draw [<->,line width=0.5pt] (-4.684168628479766,3.5344244009624575)-- (-3.111842916637063,3.533177106778101); \draw (-4.824098716609174,4.355859061130377) node[anchor=north west] {\tiny Emission Rad.}; \draw [->,>=latex,line width=0.5pt,color=wrwrwr] (-3.851360362859914,4.069178644265877) -- (-3.851360362859914,3.586274357873309); \draw[->,line width=0.6pt, color=wrwrwr](-5.472612873213186,1.9292776215477105).. controls (-5.172612873213186,1.7) and (-4.472612873213186,1.7) ..(-4.170447268187056,1.4255281261130782); \draw[->,line width=0.6pt, color=wrwrwr](-1.010277758642073,2.753579528894802).. controls (-1.010277758642073,2.053579528894802) and (-2.010277758642073,2.153579528894802) ..(-2.9447268187056,1.5255281261130782); \draw [fill=rvwvcq] (-1.9127272727272742,3.5345454545454564) circle (2.5pt); \draw[color=rvwvcq] (-1.8582488932305357,4.002643534747938) node {Emitted Defect}; \draw [fill=rvwvcq] (-6.749090909090913,3.534545454545457) circle (4.5pt); \draw[color=rvwvcq] (-6.663558737816434,4.057249328436415) node {Parent Defect}; \end{tikzpicture} $$
shift_cntr@
Only used when simulating annealing of single cascades.
cascade_dep_rate@
- Required for NEUTRON-IRRADIATION Simulations.
- This keyword is used along with
read_cascade.
read_cascade@
Usage: read_cascade <number of files> <filename_prefix>
Example
read_cascade 15 cascade- a cascade is randomly picked and read from one of the 15 cascades files with filenames cascade-1,cascade-2,…., cascade-15.
Ignored when the cascade_dep_rate is zero.
list_size@
Increases the size of the defect matrix that stores information on defect clusters.
Space for the defect matrix is dynamically allocated. The idea is to allocate the memory in larger chunks as the number of defect clusters increases during simulation, reducing the frequency of dynamic memory allocations. Memory reallocation is a computationally expensive operation..
Example
list_size 100 increases the size of the defect matrix by 100. That is, if the number of defects read from an initial cascade is 30, then the size of the defect matrix would be 130 instead of being 30.
max_walltime@
max_r_itr@
Reaction events are carried out until no reaction events are possible. This process is executed recursively and max_r_itr is used to specify the maximum number of recursive iterations allowed to carry out reaction events.
Warning
- This option should ONLY be used for debugging the code
- Best to use the default value
- Usually, it should not take more than
2-3recursive iterations to execute all possible reaction events after each KMC step.
dumpconfig@
Usage: dumpconfig <Num. of snaps> <filename_prefix>
Example
dumpconfig 10 configs_saves10snap shots of a simulation with filenames starting withconfigs_.i.e, snap shot are saved in filesconfigs_1,configs_2....configs_10dumpconfig 10 *saves10snap shots with filenames starting withsnap_shot_, i.e., snap shots are saved in filessnap_shot_1,snap_shot_2..snap_shot_10.snap_shot_is the default filename prefix
rate_scaling@
Keyword: rate_scaling is a non-essential keyword to specify the values of \(\alpha\). Default value of \(\alpha=0\)
- Diffusion rate of mobile defects can be scaled as \((size)^{-\alpha}\)
- In the input script file, values of \(\alpha\) for each defect type should be entered.